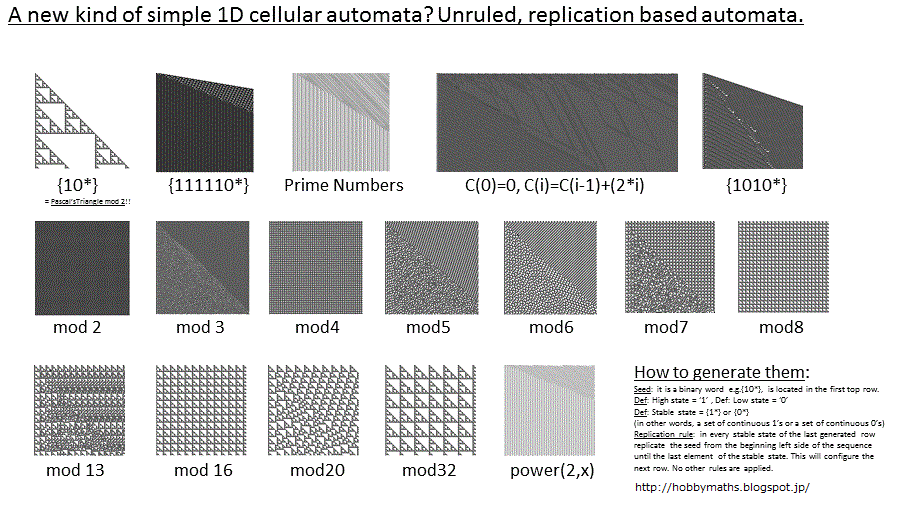
I came across a way of creating very simple 1D cellular automata, that might be new, still I did not find any references about this method in the main sources of information about this kind of structures.
The type I have been studying is a little bit different from the ones explored and extended by Stephen Wolfram (previously by Von Neumann, Ulam...), but that could be included on his classification. I call them "self-replication cellular automata". It is not a substitution system, neither a cellular automaton, and according to Wolfram's classification it is a one-dimensional neighbour dependant structure.
I think is in the middle of them, sharing properties of both types of pattern generation types. Initially I did not find any reference inside the Wolfram's automata atlas, and other pattern generation types classifications, so it could be possible that my so-called "self-replication" automaton is still not classified with samples per se.
The results in terms of the complexity of the behavior of a self-replication automaton when it evolves are very similar to the ones obtained by using the standard replication rules classified by Wolfram. (e.g. Rule 30, Rule 90, etc.) but without using them.
It is simpler, so all the rich complexity that can be obtained by using the more elaborated standard replication rules is not available in all its full extent by using my self-replication version, but is very close to some of the behaviors of the standard structures and closer to what I think could be found in real life.
The replication rule is as follows.
1. The seed: or original state of the 1D cellular automaton, or first row of the evolution graphic, is a sequence or word, for instance {10*} (= "1000000..."), or whatever sequence we want to study, for instance "mod 2" is the sequence in which we put a high value '1' to the points that are positioned in those numbers of the sequence that are multiple of 2, including the position 0 as the first element of the sequence. This is the seed for that case: {10101010...} = {(10)*} We can study any sequence, e.g. prime numbers, perfect squares, arithmetic series, etc.
2. A "High state" will be defined as a continuous sequence of '1's and a "Low state" will be a continuous sequence of '0's. A stable state is defined as a "High state" or a "Low state", independently of the quantity of elements of the high or low state (an isolated 0 or an isolated 1 is also a stable state).
3. The next evolution of the automaton will be performed as follows: from left to right, in the last generated sequence of the automaton (in the first step it is the seed) the high states {1*} are replaced with the seed again (form left to right) and same for low states. That is all.
And here are some results (click to enlarge).
It is possible to reproduce for instance the famous Sierpinski gasket (Pascal's triangle mod 2) by using the seed {10*}, and also some other sequences to visualize turbulence in boundaries, as the ones generated by the standard Wolfram's rules. The main visual difference is that avoiding the substitution rules makes the orientation of the self-replication patterns a little bit different from the standard cellular automaton (which is usually symmetrical) and the results are not so wide as the ones obtained by the use of the standard rules, it is a reduced set of those results.
It is possible also to use more that 2 states (1,0), for instance replacing in the seed stable states with a natural number sequence {0123...} and using the same replication rule it is possible to obtain a very complex cellular automaton with (in)finite states. Applying a different color to a different value provides very interesting patterns (for instance replacing the {0,1,2,3..} values of the sequence by a black-to-red-to-yellow-to-white gradient depending on the growing values shows a nice visual fractal-like pattern).
The main advantage about the self-replication patterns is that they are simpler than the rule-generated automaton, and (I guess) closer to the ones that could be biologically generated. For instance, always mollusc shells are shown as a sample of this kind of patterns, but being dependant on a rule (e.g. the above mentioned Rule 30) for a replication of the patters seems more complex than a replication simply based on two different states (high, low) of the original pattern seed. I think that using self-replication is closer to a biological pattern, more organic, because biochemically it might be easier probably to launch a chemical reaction depending on an original chemical or electrical high or low state inside a cell or group of cells. The organism would not need "to store" genetically a more complex replication rule, it would just need to replicate the original pattern, so stable states of the pattern will lead to a replication of the original seed.
If somebody already heard about this kind of more simple "self-replication" automaton patterns I found please let me know, I would like to learn more about them if they were defined before.